- People
- Research
- Graduate
-
Undergraduate
- Prospective & Transfer Students
- General Chemistry Options
- Policies
- Tutors
- Pre-Health
- Undergraduate Advisors
-
Chemistry Major
- Process for Declaring a Major, Minor, DMP, or ACS Certification
- B.A. in Chemistry
- B.S. Chemistry
- B.S. Specialization in Biochemistry
- B.S. Specialization in Chemical Education
- B.S. Specialization in Chemical Physics
- B.S. Specialization in Environmental Chemistry
- B.S. Specialization in Materials Science
- B.A./M.S. or B.S./M.S. in Chemistry ("3+1" Degree Option)
- Undergraduate Research
- Distinguished Majors Program
- Minor
- Forms
- Study Abroad
- Undergraduate FAQs
- Undergraduate Resources
- Safety
- Seminars
- Newsletter

The department has four EPR spectrometers with multifrequency and pulsed capabilities.

The Department of Chemistry has two Hi-Resolution Mass Spectrometers.

The nanoSTAR Institute encompasses nanoscale and quantum research.

The UVa NMR Spectroscopy Core Facility has five spectrometers for solution samples.

The stockroom is located in room 221 of the Chemistry Building, at the loading dock on the second floor, and carries most common chemical and supplies for the teaching and research laboratories.

Designed to simplify the process of finding local research technology solutions.
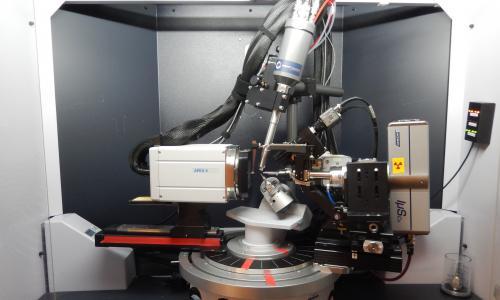
This facility is equipped with multiple X-ray diffractometers for single-crystal structural determination of small molecules and macromolecules, as well as powder diffraction studies.